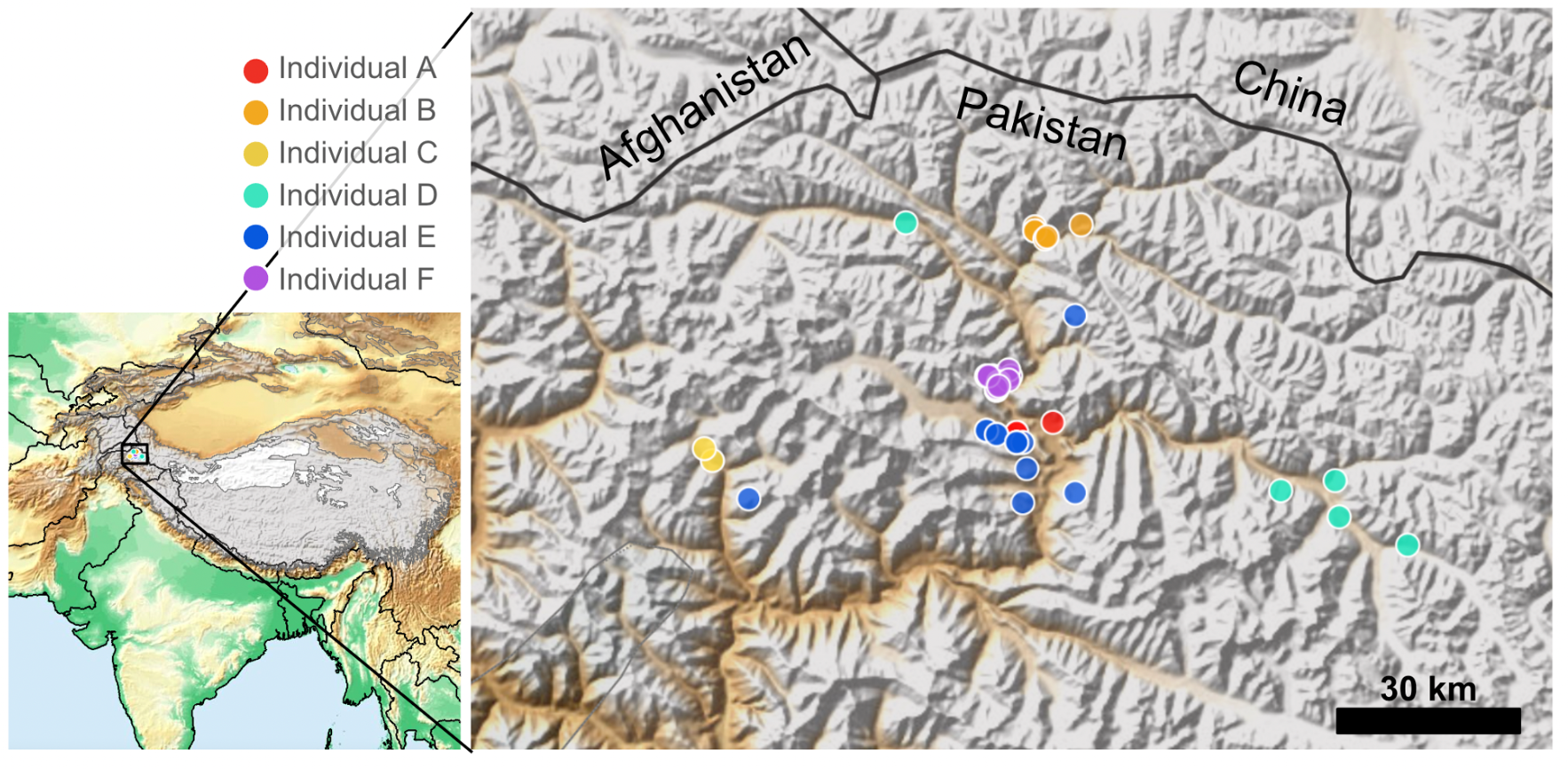
SLN invites you for our next webinar about a community-based snow leopard monitoring system in Nepal’s Nar and Phu Valley, integrating mobile-based tools for data collection and conflict mitigation. (Snow Leopard Reports, 4 (2025): 1-14)

About the Talk:
Snow leopards live in some of the most remote mountain regions in the world. But their biggest threat is often conflict with people. When livestock are killed, families can lose a significant part of their income. This creates tension and sometimes leads to retaliation.
In this webinar, Arun will share how they worked with local communities in Nepal’s Nar–Phu Valley to develop a simple mobile-based monitoring system. Together with herders and local conservation committees, they co-designed a smartphone app that allows community members to record snow leopard signs, register livestock, and report livestock losses — even in areas with limited internet access.
Between 2023 and 2024, community members recorded 483 snow leopard observations and reported depredation cases in a structured way that supports compensation and prevention efforts. The project also helped identify vulnerable livestock corrals and led to the construction of predator-proof enclosures.
This session will focus on practical lessons: how to design tools that people actually use, how to build trust and ownership, and how mobile technology can strengthen coexistence between wildlife and communities. The approach is simple, adaptable, and relevant not only for snow leopards, but for many human–wildlife conflict situations worldwide.


About our Speaker:
Dr. Arun Kumar Pratihast is a Senior Researcher at Wageningen Environmental Research (Wageningen University & Research, The Netherlands). He has more than 15 years of experience in community-based monitoring, citizen science, and the use of mobile and geospatial technologies for conservation and sustainable land management. Over the years, he has worked closely with local communities, NGOs, and governments across Asia, Africa, and Latin America to design practical monitoring systems that are simple, scalable, and built around local needs. His work focuses on combining technology with local knowledge to support better decisions for biodiversity and people.


About our Facilitator:
Joshua Schlüter is a conservationist working with the Nature And Biodiversity Conservation Union (NABU e.V.) based in Germany. His work focuses on species conservation in Central Asia and Nepal, with a particular emphasis on mitigating human–wildlife conflicts. In the region, NABU supports long-term snow leopard conservation through community-based approaches, research, and cross-border cooperation. Joshua implements projects in close collaboration with local partners, aiming to strengthen local capacities and achieve long-term conservation impact. More recently, he has also expanded his work to include engagement with international environmental policy processes under conventions such as CITES and CMS
Date/Time:
Thursday, 5th March at 15:00 PM (Bishkek time)
Location:
ZOOM, to join this talk, REGISTER HERE
Please note:
- If you have never used Zoom before, we recommend that you try the link 10 minutes before the start of the lecture.
- Please feel free to write questions in the comment area and there will be time for questions/discussion at the end of the talk.
- Please note that the session will be recorded and later featured on the SLN website. If you have concerns about this please let us know before the session