|
|
Johnson, W. E., Dratch, P. A., Martenson, J. S., & O'Brien, S. J. (1996). Resolution of recent radiations within three evolutionary lineages of Felidae using mitochondrial restriction fragment length polymorphism variation. Journal of Mammalian Evolution, 3(2), 97–120.

Abstract: Patterns of mitochondrial restriction fragment length polymorphism (RFLP) variation were used to resolve more recent relationships among the species of the Felidae ocelot lineage, domestic cat lineage, and pantherine lineage. Twenty-five of 28 restriction enzymes revealed site variation in at least 1 of 21 cat species. The ocelot lineage was resolved into three separate sister taxa groups: Geoffroy's cat (Oncifelis geoffroyi) and kodkod (O. guigna), ocelot (Leopardus pardalis) and margay (L. wiedii), and pampas cat (Lynchailurus colocolo) and most of the tigrina samples (Leopardus tigrina). Within the domestic cat lineage, domestic cat (Felis catus), European wild cat (F. silvestris), and African wild cat (F. libyca) formed a monophyletic trichotomy, which was joined with sand cat (F. margarita) to a common ancestor. Jungle cat (F. chaus) and black-footed cat (F. nigripes) mtDNAs diverged earlier than those of the other domestic cat lineage species and are less closely related. Within the pantherine lineage, phylogenetic analysis identified two distinct groups, uniting lion (P. leo) with leopard (P. pardus) and tiger (P. tigris) with snow leopard (P. uncia).
|

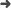
|
|
|
Johnson, W. E., Eizirik, E., Pecon-Slattery, J., Murphy, W. J., Antunes, A., Teeling, E., et al. (2006). The Late Miocene Radiation of Modern Felidae: A Genetic Assessment (Vol. 311).

Abstract: Modern felid species descend from relatively recent (G11 million years ago) divergence and speciation events that produced successful predatory carnivores worldwide but that have confounded taxonomic classifications. A highly resolved molecular phylogeny with divergence dates for all living cat species, derived from autosomal, X-linked, Y-linked, and mitochondrial gene segments (22,789 base pairs) and 16 fossil calibrations define eight principal lineages produced through at least 10 intercontinental migrations facilitated by sea-level fluctuations. A ghost lineage analysis indicates that available felid fossils underestimate (i.e., unrepresented basal branch length) first occurrence by an average of 76%, revealing a low representation of felid lineages in paleontological remains. The phylogenetic performance of distinct gene classes showed that Y-chromosome segments are appreciably more informative than mitochondrial DNA, X-linked, or autosomal genes in resolving the rapid Felidae species radiation.
|

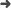
|

